Molecular Modeling
Structural modeling of proteins and protein-ligand complexes
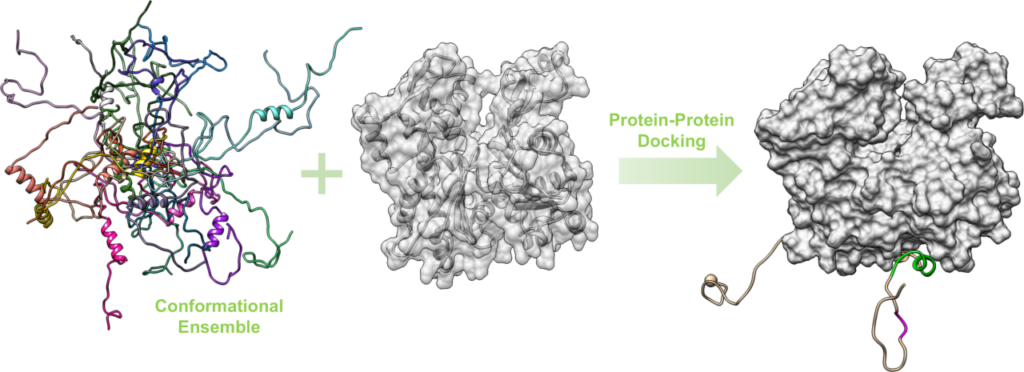
Structure-based drug design is the main strategy used in our molecular modeling group to support the research and development of new therapeutic compounds by the BioCIS medicinal chemists. Thus, one of our major activities is to investigate the three-dimensional structures of drug targets which are mainly proteins and protein-protein interactions (PPIs). We are particularly interested in intrinsically disordered proteins and their complexes which conformational ensembles are often difficult to be characterized experimentally. For this purpose, we generally use classical or enhanced molecular dynamics simulations, as well as protein-protein docking calculations. Our studies include :
- Structural ensemble and biological activity of DciA intrinsically disordered region. Chan-Yao-Chong M, Marsin S, Quevillon-Cheruel S, Durand D, Ha-Duong T. J Struct Biol. 2020 ; 212 : 107573.
- Investigation into Early Steps of Actin Recognition by the Intrinsically Disordered N-WASP Domain V. Chan-Yao-Chong M, Durand D, Ha-Duong T. Int J Mol Sci. 2019 ; 20 : 4493.
- Structural Characterization of N-WASP Domain V Using MD Simulations with NMR and SAXS Data. Chan-Yao-Chong M, Deville C, Pinet L, van Heijenoort C, Durand D, Ha-Duong T. Biophys J. 2019 ; 116 : 1216-1227.
- Conformational Ensemble and Biological Role of the TCTP Intrinsically Disordered Region : Influence of Calcium and Phosphorylation. Malard F, Assrir N, Alami M, Messaoudi S, Lescop E, Ha-Duong T. J Mol Biol. 2018 ; 430 : 1621-1639.
- Interaction of chemokine receptor CXCR4 in monomeric and dimeric state with its endogenous ligand CXCL12 : coarse-grained simulations identify differences. Cutolo P, Basdevant N, Bernadat G, Bachelerie F, Ha-Duong T. J Biomol Struct Dyn. 2017 ; 35 : 399-412.
- Effect of Post-Translational Amidation on Islet Amyloid Polypeptide Conformational Ensemble : Implications for Its Aggregation Early Steps. Tran L, Ha-Duong T. Int J Mol Sci. 2016 ; 17 : 1896.
- Structure of ring-shaped Aβ₄₂ oligomers determined by conformational selection. Tran L, Basdevant N, Prévost C, Ha-Duong T. Sci Rep. 2016 ; 6 : 21429.
- Comparative study of structural models of Leishmania donovani and human GDP-mannose pyrophosphorylases. Daligaux P, Bernadat G, Tran L, Cavé C, Loiseau PM, Pomel S, Ha-Duong T. Eur J Med Chem. 2016 ; 107 : 109-18.
Self-assembly of biopolymers for encapsulation of bioactive molecules
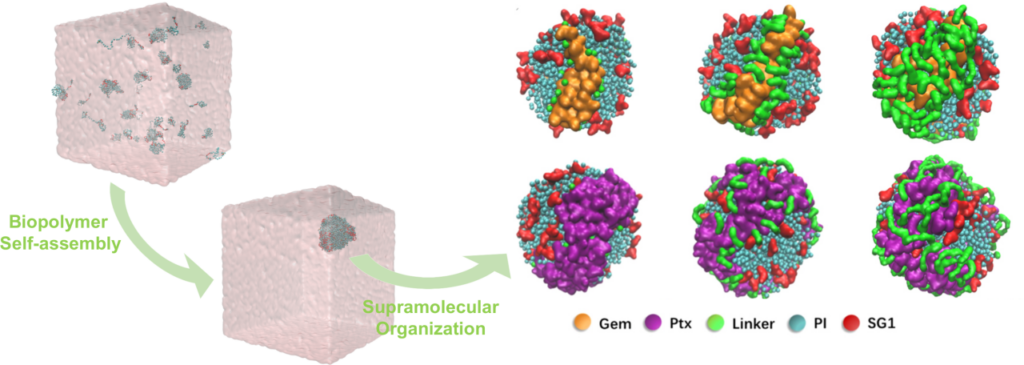
Related to our interest in intrinsically disordered proteins, our group has established strong collaborations with Institut Galien Paris-Saclay to help the development of biopolymers for efficient drug loading into nanocarriers and controlled release in targeted tissues. By using advanced coarse-grained molecular dynamics simulations, we study the self-assembly process and the liquid-liquid phase separation of thermo-sensitive biopolymers and investigate the supramolecular organization of drug polymer nanoparticles. The aim of these recent studies is to gain insight into the physico-chemical and structural properties that govern the formation and degradation of these biopolymer nanoparticles. Here are reports of our recent results :
- Supramolecular Organization of Polymer Prodrug Nanoparticles Revealed by Coarse-Grained Simulations. Gao P, Nicolas J, Ha-Duong T. J Am Chem Soc. 2021 ; 143 : 17412-17423.
- Simulations of the Upper Critical Solution Temperature Behavior of Poly(ornithine-co-citrulline)s Using MARTINI-Based Coarse-Grained Force Fields. Molza AE, Gao P, Jakpou J, Nicolas J, Tsapis N, Ha-Duong T. J Chem Theory Comput. 2021 ; 17 : 4499-4511.
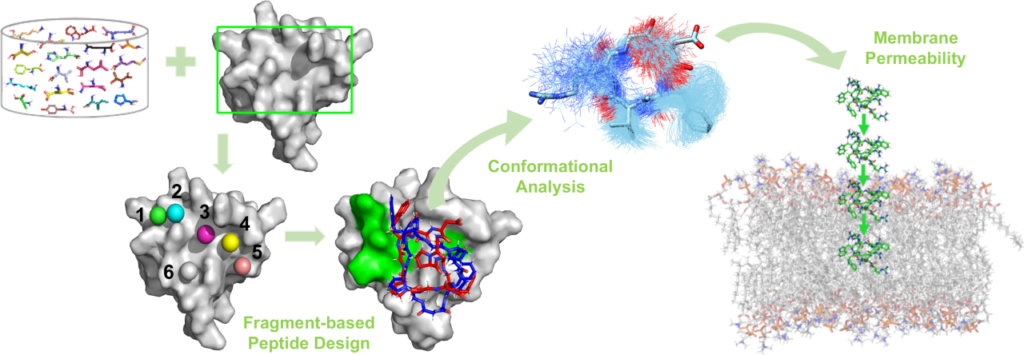
Design of peptidomimetic inhibitors of protein-protein interactions
As protein-protein interactions (PPIs) are involved in many physiopathological processes, searching for PPIs inhibitors is a promising but challenging strategy in drug design. As part of the FLUOPEPIT team, the molecular modeling group develops and applies computational methods to design and study peptides and peptidomimetics that (i) would likely bind a targeted protein-protein interface with high affinity and (ii) would have good proteolytic stability and membrane permeability. Our strategy for this purpose is a fragment-based approach that identify the optimal sequences of cyclic peptides for binding a specific protein surface. Furthermore, we use enhanced molecular dynamics simulations to characterize conformational ensembles of various (cyclic, fluorinated, aza… ) peptidomimetics and their membrane permeability. Our contributions in the peptide field include :
- Understanding Passive Membrane Permeation of Peptides : Physical Models and Sampling Methods Compared. Mazzanti L, Ha-Duong T. Int J Mol Sci. 2023 ; 24 : 5021.
- Computational design of cyclic peptides to inhibit protein-peptide interactions. Delaunay M, Ha-Duong T. Biophys Chem. 2023 ; 296 : 106987.
- Des3PI : a fragment-based approach to design cyclic peptides targeting protein-protein interactions. Delaunay M, Ha-Duong T. J Comput Aided Mol Des. 2022 ; 36 : 605-621.
- β-Hairpin Peptide Mimics Decrease Human Islet Amyloid Polypeptide (hIAPP) Aggregation. Lesma J, Bizet F, Berardet C, Tonali N, Pellegrino S, Taverna M, Khemtemourian L, Soulier JL, van Heijenoort C, Halgand F, Ha-Duong T, Kaffy J, Ongeri S. Front Cell Dev Biol. 2021 ; 9 : 729001.
- Fluorinated Triazole Foldamers : Folded or Extended Conformational Preferences. Laxio-Arenas J, Xu Y, Milcent T, Van Heijenoort C, Giraud F, Ha-Duong T, Crousse B, Ongeri S. ChemPlusChem. 2021 ; 86 : 241-251.
- Binding Modes of a Glycopeptidomimetic Molecule on Aβ Protofibrils : Implication for Its Inhibition Mechanism. Tran L, Kaffy J, Ongeri S, Ha-Duong T. ACS Chem Neurosci. 2018 ; 9 : 2859-2869.
- Structure-activity relationships of β-hairpin mimics as modulators of amyloid β-peptide aggregation. Tonali N, Kaffy J, Soulier JL, Gelmi ML, Erba E, Taverna M, van Heijenoort C, Ha-Duong T, Ongeri S. Eur J Med Chem. 2018 ; 154 : 280-293.
- The use of 4,4,4-trifluorothreonine to stabilize extended peptide structures and mimic β-strands. Xu Y, Correia I, Ha-Duong T, Kihal N, Soulier JL, Kaffy J, Crousse B, Lequin O, Ongeri S. Beilstein J Org Chem. 2017 ; 13 : 2842-2853.